by Mohamed Masoud, Farfalla Hu, and Sergey Plis
Introduction
Extracting brain tissue from MRI volumes and segmenting it either into gray and white matter or into more elaborate brain atlas is an essential step in many brain imaging analysis pipelines. Surgical planning, measuring brain changes and visualizing its anatomical structures are just a few of more clinical applications commonly dependent on MRI segmentation. In this post, we present brainchop.org that brings automatic MRI segmentation capability to neuroimaging by running a robustly pre-trained deep learning model in a web-browser on the user side. brainchop.org is a client-side web-application for automatic segmentation of MRI volumes. The app does not require technical sophistication from the user and is designed for locally and privately segmenting user’s T1 volumes. Results of the segmentation may be easily saved locally after the computation. An intuitive interactive interface that does not require any special training nor specific instruction to run enables access to a state of the art deep learning brain segmentation for anyone with a modern browser (e.g. Firefox, Chrome etc) and commonly available hardware. Additionally, we make implementation of brainchop.org freely available releasing its pure javascript code as open-source. The user interface (UI) provides a web-based end-to-end solution for 3D MRI segmentation as shown in Figure 1.
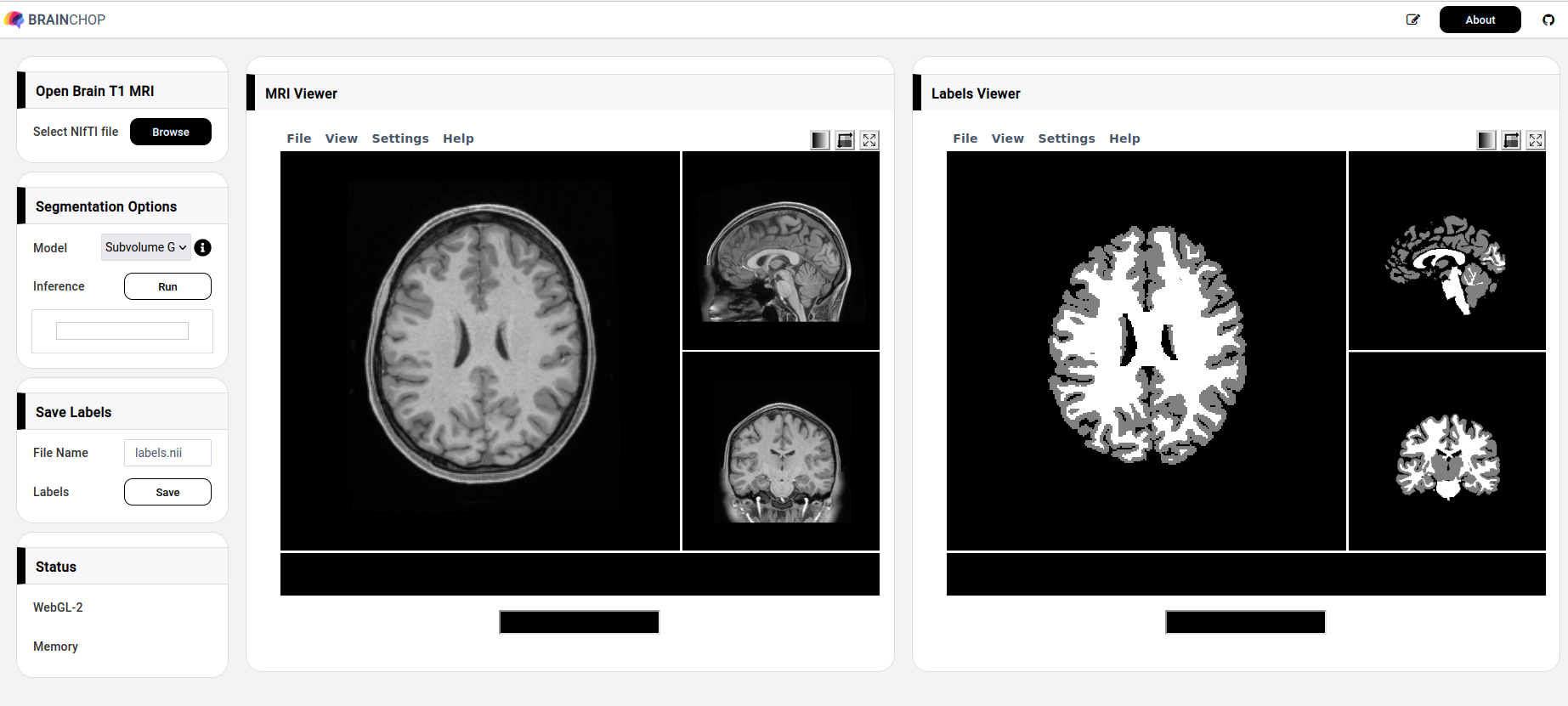
You are welcome to explore brainchop.org before digging further into the tool details. Notably, despite the need to load a few fully trained models, brainchop.org opens very quickly as the traffic is negligible, thanks to the parsimonious architecture of our models. Continue reading to learn the details.
Motivation
For many researchers and radiologists, especially in developing countries, setting up neuroimaging pipelines is a technological barrier and providing those pipelines through the browser will help democratize these computational approaches. Existing solutions are either difficult to set up locally, or require data transfer offsite (cloud, or resource provider servers), which is not always possible or desirable. brainchop.org aims to make neuroimaging pipelines accessible from anywhere in the world while preserving the data privacy since the tool performs computation on the client-side and does not transfer the data.
Challenges with in-Browser 3D Segmentation
While using the browser for machine learning has various advantages, it is also associated with multiple challenges. One of the most important features of using web-based tools is the waiver of any software installation requirements and the high accessibility of the tool online. However, to maintain the easy-to-use advantage with frontend data science tools, numerous difficulties have to be tackled to create a reliable and fast tool. Among these difficulties is the browser limitation in memory and resource management. Also, the shortage of publicly available machine learning and image processing javascript libraries makes creating such applications very time consuming and may lead to development of all necessary libraries from scratch.
Problem Definition
Given a 3D MRI brain volume, the task is to segment brain tissue within a browser on the user side into regions of interest such as the Gray-Matter and White Matter (GMWM) regions. In many cases, such a task requires a pre-trained lightweight model, MeshNet or similar, to simply be able to run in the browser. The fewer parameters a model has, the better for the browser performance. Yet, there is a tradeoff between the model size and its accurate performance. We could, for example, make the model smaller by making it work on flat slices of the volume. This, however, would be detrimental to the model accuracy as a side effect. It’s been commonly realized that 3D input provides much more information to the model than 2D slices. Thus the goal is to have a model that works on volumes as its input, preferably full brain at once, is very small in size and computational requirements, and yet is highly accurate.
Data
Version 1.0.0 of Brainchop is designed to support segmentation of T1 weighted images. The only currently supported input format of those images is Nifti. For the models to work as intended, the input T1 image needs to be resampled to 1x1x1 mm thickness by using FreeSurfer command mri_convert as follows:
mri_convert original_T1.nii new_T1.nii.gz -cAfter selecting the T1 weighted image for 3D segmentation, Brainchop applies different preprocessing steps underneath such as:
– Converting data to javascript readable format.
– Converting formatted data to tensors readable by the tfjs framework.
– Partitioning tensors to 3D patches for inference when using a subvolume model.
– Image normalization to enhance model accuracy.
Full Brain vs. Subcube Models
Notably, most, if not all, earlier publications on volumetric brain segmentation both those that use U-net-based models and our own dilated convolutions MeshNet [3] work on subvolumes for memory conservation. Full brain models are only recently entering the model zoo, thanks to GPU cards with 80 GB memory [6]. Yet there is a clear reason to use a full brain model, since the segmentations are highly location specific relative to the global brain structure. To a lesser extent brain regions are determined by the local tissue structure. This is especially true for semantic segmentation for the functional atlas, however, also applies to the gray and white matter segmentation we provide in the initial version of brainchop.org. Another difference between segmentation of the full brain is that it can be done in a single forward run of the full brain model, while the subvolume model needs to run on each subvolume independently (and often sequentially) with the need to subsequently combine all results.
In the following image we have tried to convey the difference between the difficulty of segmenting the brain from subfolume and full brain segmentation.

WebGL Backend
The main goal of Brainchop is to make 3D segmentation in the browser practical, fast and accurate. Normally, T1-Images can have up to 256x256x256 dimensions, requiring careful considerations to retain efficiency of in-browser processing without memory leaks or loss of the WebGL (Web Graphics Library) context. We provide a few options to account for possible limitations of the client’s device. Most accurate and performant is the full brain model, that may not work on machines that do not have a dedicated graphics card. A smaller full brain model that is not as versatile in the types of input data it can handle and less accurate but 75% of the client-side configurations that we have tested could run it without a problem. Finally, to fully overcome browser limitations affecting full brain models we provide a model that operates by patching the 3D image data into subvolumes of tensors (e.g. 38x38x38) and cleaning up the memory for unneeded tensor allocations. Iterating through the whole MRI volume needs a frontend that utilizes a local GPU and an enabled WebGL with the browser.
Inference Model
A perfect example is the MeshNet segmentation model which has a competitive DICE score compared to the classical segmentation model U-Net and meanwhile it has much fewer parameters. We have performed a comparison between the two models’ performances on GPU, as summarized in Table-1.
| Model | Inference Speed | Model Size | Macro DICE |
| MeshNet GMWM | 116 subvolumes/sec | .89 mb | 0.96 |
| U-Net GMWM | 13 subvolumes/sec | 288 mb | 0.96 |
| MeshNet GMWM (full brain model) | 0.001 sec/volume | 0.022 mb | 0.96 |
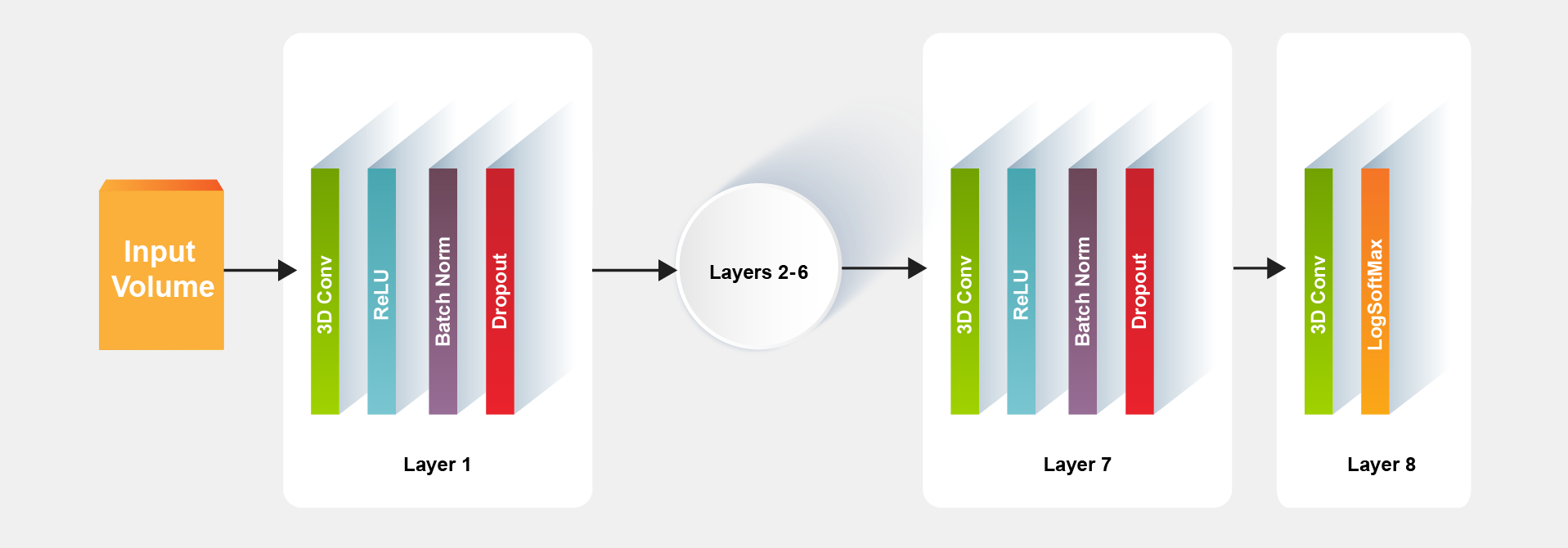
The advantage of MeshNet small size is due to its simple architecture in which a typical model for the segmentation task can be constructed with eight layers as shown in Figure 2.
Tfjs conversion
In order to deploy the MeshNet model in the browser, there is a need to convert it first to a workable tensorflow.js (tfjs) model. If the original segmentation model created with Keras, make sure your keras model layers are compatible with tfjs layers listed here.
For example, if the keras model input shape is [1, 38, 38, 38, 1], then keras batchnorm5d will be in use which will raise an issue with tfjs model because at the time of writing this blog there is not a batchnorm5d layer in tfjs. One possible workaround here is to use a fusion technique with keras layers by merging batch normalization layer with convolution layer as shown here .
After training original model on segmentation task, multiple converters to tfjs can be used from command line or by python code such as:
# Python sample code
import tensorflowjs as tfjs# Loading the saved keras model back
keras_model = keras.models.load_model('path/to/model/location')
# Convert and save keras to tfjs_target_dir
tfjs.converters.save_keras_model(keras_model, tfjs_target_dir)
For more information about importing a model e.g keras into Tensorflow.js please refer this tutorial
A sample converted tfjs model with Brainchop can be found at this link , where the folder has two main files, the model json file and the weights file.
- The JSON file consists of modelTopology and weightsManifest.
- The binary weights file consists of the concatenated weight values.
Optionally another two json files can be added also, the labels.json file to annotate the output labels, and the colorLUT.json file to apply customized colors to the segmentation regions.
The schema of the labels.json file can be such as:
{"0": "background", "1": "Gray Matter", "2": "White Matter"}
The object keys “0”, “1” and “2” must be numbers representing expected labels resulting from the inference model, and the values are the associated annotations for each key as shown in Fig.4.
For colorLUT.json file, it is another optional file with the following schema:
{"0": "rgb(0,0,0)", "1": "rgb(0,255,0)", "2": "rgb(0,0,255)"}
Where the json keys must be numbers represent the segmentation labels, and the value represents the color associated with that label in rgb format. Loading above files can be done using the model browsing option from model list as shown in Figure 5.
Brainchop Post Processing
After the inference is complete, if the subvolume model was used Brainchop rebuilds the complete 3D MRI volume from model output patches that cover the entire volume as a grid. We can also add overlapping randomly sampled subvolumes to increase accuracy in exchange for a small slowdown. The full brain models perform a single forward pass in this case. 3D noisy clusters outside the brain may result from the inference process due to possible bias, variance and irreducible error (e.g. noise in the data). To remove these noisy clusters we designed a 3D connected components algorithm to filter out those noisy clusters.
To visualize the input and output images we used papaya viewers. A composite operation is also provided to subjectively verify the output image accuracy. Different ColorLUT can be applied to enhance the visualization process. After verifying the output labels the segmentation can be saved locally as a Nifti file.

Brainchop Performance
With an average GPU of 768 cores/ 4GB buffer, 7Gbps memory speed and a system memory of 16 GB, Brainchop shows fast response while processing 256x256x256 Nifty images in around 30 seconds. For the subvolume model, the inference iterates through 64 subcubes that cover the T1 each of shape 64x64x64. In this case, the pipeline takes longer, usually around 35-40 seconds to complete the whole task. We have tested this on multiple browsers that support WebGL-2 and multiple hardware configurations.
Next addons
Our future work on Brainchop v2.0.0 will include segmenting into functional atlases. We also consider a possible inclusion of models that are insensitive to voxel size of the input and, potentially, to other imaging parameters. This could remove the need of pre-processing with mri_convert.
Conclusions
Despite the challenges associated with deploying large machine learning models in the browsers, the attempts are still worthy and promising. In this post, we presented a strong proof-of-concept of the browser’s capability to quickly run volumetric brain segmentation. With an appropriate inductive bias a model that is powerful and simultaneously economical in size and computation can find a great use for in-browser client-side applications. Our MeshNet implementation is solid evidence of this. The accessibility, scalability, ease of use, lack of installation requirements and cross-platform operation are just a few of the unique and enabling features that a web-based in-browser application can provide while also preserving end-user data privacy.
FInally, We welcome your feedback to help shape our priorities for further development of brainchop.org. We also welcome contributors familiar with tfjs and neuroimaging applications (e.g. tumor segmentation, etc) who are interested in getting involved in expanding brainchop.org model zoo.
Looking forward to your contribution!
Acknowledgements
This work was supported by the NIH grant RF1MH121885.
We thank Alex Fedorov for MeshNet, Kevin Wang for his great help in training models and running experiments, and Sergey Kolesnikov for Catalyst and work on Catalyst.Neuro.
References:
- Brainchop github repository: https://github.com/neuroneural/brainchop
- Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift https://arxiv.org/abs/1502.03167
- F. Yu and V. Koltun, “Multi-scale context aggregation by dilated convolutions,” arXiv preprint arXiv:1511.07122, 2015
- Alex Fedorov, Jeremy Johnson, Eswar Damaraju, Alexei Ozerin, Vince Calhoun, and Sergey Plis. End-to-end learning of brain tissue segmentation from imperfect labeling. In 2017 IEEE International Joint Conference on Neural Networks (IJCNN), 3785-3792, 2017.
- Catalyst.Neuro: A 3D Brain Segmentation Pipeline for MRI using Catalyst
- Partial Volume Segmentation of Brain MRI Scans of any Resolution and Contrast B. Billot, E.D. Robinson, A.V. Dalca, J.E. Iglesias MICCAI 2020
- Papaya : https://github.com/rii-mango/Papaya
- NIfTI Reader: https://github.com/rii-mango/NIFTI-Reader-JS
- TensorFlow.js : https://www.tensorflow.org/js
